티스토리 뷰
1. phenofit 사이트
https://phenofit.top/index.html
Extract Remote Sensing Vegetation Phenology
The merits of TIMESAT and phenopix are adopted. Besides, a simple and growing season dividing method and a practical snow elimination method based on Whittaker were proposed. 7 curve fitting methods and 4 phenology extraction methods were provided. Paramet
phenofit.top
2. 필요한 패키지 설치전 버전 확인
1. R 버전 : [64-bit] C:\Program Files\R\R-4.2.3
2. R studio 버전:
3. phenofit 3.0 버전 이용 : https://github.com/eco-hydro/phenofit-scripts
GitHub - eco-hydro/phenofit-scripts: phenofit examples in version 3.0
phenofit examples in version 3.0. Contribute to eco-hydro/phenofit-scripts development by creating an account on GitHub.
github.com
3. 관련 패키지 설치
phenofit 버전 3.0을 사용하기 위해서 사전에 설치되어야 한 패키지 설치
# install.packages("remotes")
remotes::install_github("eco-hydro/phenofit")
or
library(remotes)
install_github("eco-hydro/phenofit")
install_github("rpkgs/Ipaper")
install_github("rpkgs/sf.extra")
install_github("rpkgs/lattice.layers")
4. 기본예제
1) 패키지 로드
library(phenofit)
library(data.table)
library(dplyr)
library(ggplot2)
2) 자료 로드
data(MOD13A1)
d = MOD13A1$dt %>% subset(site == "CA-NS6" & date >= "2010-01-01" & date <= "2016-12-31") %>%
.[, .(date, y = EVI/1e4, DayOfYear, QC = SummaryQA)]
d %<>% mutate(t = getRealDate(date, DayOfYear)) %>%
cbind(d[, as.list(qc_summary(QC, wmin = 0.2, wmid = 0.5, wmax = 0.8))]) %>%
.[, .(date, t, y, QC_flag, w)]
print(d)
#> date t y QC_flag w
#> 1: 2010-01-01 2010-01-08 0.1531 snow 0.2
#> 2: 2010-01-17 2010-01-25 0.1196 snow 0.2
#> 3: 2010-02-02 2010-02-13 0.1637 snow 0.2
#> 4: 2010-02-18 2010-02-25 0.1301 snow 0.2
#> 5: 2010-03-06 2010-03-21 0.1076 snow 0.2
#> ---
#> 157: 2016-10-15 2016-10-23 0.1272 snow 0.2
#> 158: 2016-10-31 2016-11-07 0.1773 snow 0.2
#> 159: 2016-11-16 2016-11-25 0.0711 cloud 0.2
#> 160: 2016-12-02 2016-12-12 0.1372 snow 0.2
#> 161: 2016-12-18 2017-01-02 0.1075 snow 0.2
3) phenofit 초기 변수값
lambda <- 8
nptperyear <- 23
minExtendMonth <- 0.5
maxExtendMonth <- 1
minPercValid <- 0
wFUN <- wTSM # wBisquare
wmin <- 0.2
methods_fine <- c("AG", "Zhang", "Beck", "Elmore", "Gu")
4) 생물계절 변화 시기 구분 및 미세 곡선 피팅
단순히 달력상의 연도를 완전한 생육 시즌으로 간주하면 phenofit 추출에 상당한 오류가 발생할 수 있습니다. phenofit 에서는 간단한 생물계절 변화 시기 구분 방법을 제안했습니다. 생물계절 변화 시기 구분 방법은 휘태커 스무더에 크게 의존합니다. 초기 가중치, 생육기 분할, 곡선 피팅, 페노로지 추출의 절차가 별도로 수행됩니다.
INPUT <- check_input(d$t, d$y, d$w,
QC_flag = d$QC_flag,
nptperyear = nptperyear,
maxgap = nptperyear / 4, wmin = 0.2
)
brks <- season_mov(INPUT,
list(FUN = "smooth_wWHIT", wFUN = wFUN,
maxExtendMonth = 3,
wmin = wmin, r_min = 0.1
))
# plot_season(INPUT, brks)
## 2.4 Curve fitting
fit <- curvefits(INPUT, brks,
list(
methods = methods_fine, # ,"klos",, 'Gu'
wFUN = wFUN,
iters = 2,
wmin = wmin,
# constrain = FALSE,
nextend = 2,
maxExtendMonth = maxExtendMonth, minExtendMonth = minExtendMonth,
minPercValid = minPercValid
))
## check the curve fitting parameters
l_param <- get_param(fit)
print(l_param$Beck)
#> # A tibble: 7 × 7
#> flag mn mx sos rsp eos rau
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2010_1 0.191 0.538 3808. 0.0826 3894. 0.0432
#> 2 2011_1 0.194 0.477 4180. 0.113 4274. 0.113
#> 3 2012_1 0.189 0.514 4542. 0.196 4643. 0.0864
#> 4 2013_1 0.190 0.493 4903. 0.190 5011. 0.0672
#> 5 2014_1 0.198 0.494 5276. 0.122 5375. 0.289
#> 6 2015_1 0.212 0.493 5639. 0.142 5726. 0.169
#> 7 2016_1 0.198 0.501 6002. 0.186 6095. 0.0731
dfit <- get_fitting(fit)
print(dfit)
#> flag t y QC_flag meth ziter1 ziter2
#> 1: 2010_1 2010-01-25 0.1196 snow AG 0.1888598 0.1903677
#> 2: 2010_1 2010-01-25 0.1196 snow Zhang 0.1881071 0.1897593
#> 3: 2010_1 2010-01-25 0.1196 snow Beck 0.1888971 0.1906256
#> 4: 2010_1 2010-01-25 0.1196 snow Elmore 0.1882156 0.1906010
#> 5: 2010_1 2010-01-25 0.1196 snow Gu 0.1876635 0.1878432
#> ---
#> 761: 2016_1 2016-12-12 0.1372 snow AG 0.1933491 0.1963926
#> 762: 2016_1 2016-12-12 0.1372 snow Zhang 0.1939335 0.1981793
#> 763: 2016_1 2016-12-12 0.1372 snow Beck 0.1940403 0.1985395
#> 764: 2016_1 2016-12-12 0.1372 snow Elmore 0.1941100 0.1986073
#> 765: 2016_1 2016-12-12 0.1372 snow Gu 0.1872110 0.1872110
## 2.5 Extract phenology
TRS <- c(0.1, 0.2, 0.5)
l_pheno <- get_pheno(fit, TRS = TRS, IsPlot = FALSE) # %>% map(~melt_list(., "meth"))
print(l_pheno$doy$Beck)
#> flag origin TRS1.sos TRS1.eos TRS2.sos TRS2.eos TRS5.sos TRS5.eos
#> 1: 2010_1 2010-01-01 126 300 137 280 154 248
#> 2: 2011_1 2011-01-01 143 278 151 270 163 257
#> 3: 2012_1 2012-01-01 148 288 153 277 160 261
#> 4: 2013_1 2013-01-01 142 298 147 284 154 262
#> 5: 2014_1 2014-01-01 143 271 151 267 163 262
#> 6: 2015_1 2015-01-01 144 262 150 256 161 248
#> 7: 2016_1 2016-01-01 147 285 151 272 159 253
#> DER.sos DER.pos DER.eos UD SD DD RD Greenup Maturity Senescence Dormancy
#> 1: 155 192 242 132 175 211 289 128 184 213 295
#> 2: 163 210 257 146 181 240 277 142 185 236 279
#> 3: 160 194 261 150 170 239 284 145 175 233 288
#> 4: 155 187 263 144 165 236 293 139 170 228 297
#> 5: 163 230 262 146 179 253 273 142 183 250 274
#> 6: 161 207 248 147 175 235 261 142 179 231 264
#> 7: 159 189 252 148 170 228 280 144 175 221 284
pheno <- l_pheno$doy %>% melt_list("meth")
5) 시각화
# growing season dividing
plot_season(INPUT, brks, ylab = "EVI")
# Ipaper::write_fig({ }, "Figure4_seasons.pdf", 9, 4)
# fine curvefitting
g <- plot_curvefits(dfit, brks, title = NULL, cex = 1.5, ylab = "EVI", angle = 0)
grid::grid.newpage()
grid::grid.draw(g)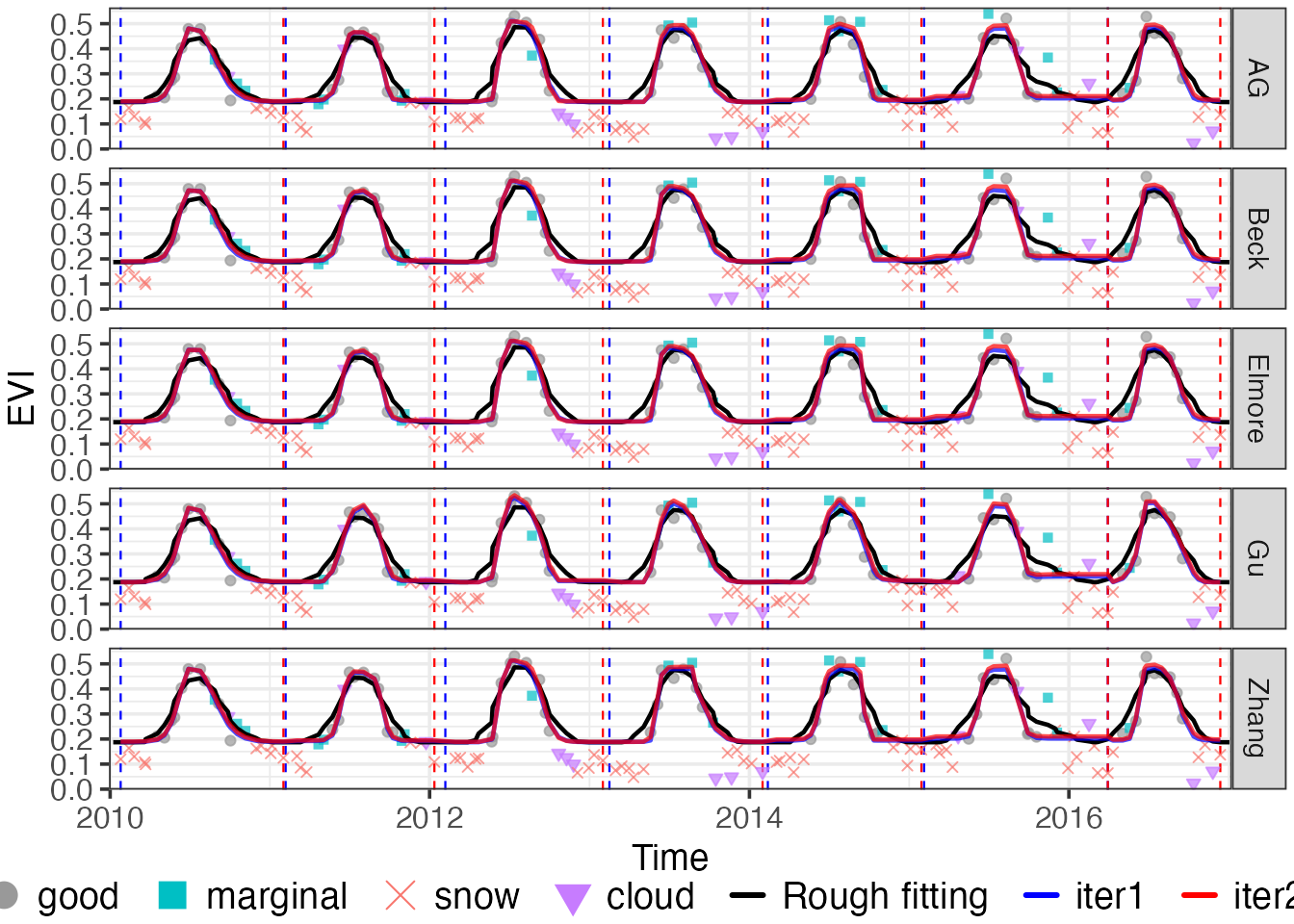
# Ipaper::write_fig(g, "Figure5_curvefitting.pdf", 8, 6, show = TRUE)
# extract phenology metrics, only the first 3 year showed at here
# write_fig({
l_pheno <- get_pheno(fit[1:7], method = "AG", TRS = TRS, IsPlot = TRUE, show.title = FALSE)
'R 통계' 카테고리의 다른 글
| 서식지 선호도 차이 여부 통계 검증 - R 카이제곱 검증 (0) | 2024.04.26 |
|---|---|
| python, R을 위한 분석을 위해 가짜 데이터 생성하기 (1) | 2024.04.26 |
